

|

|
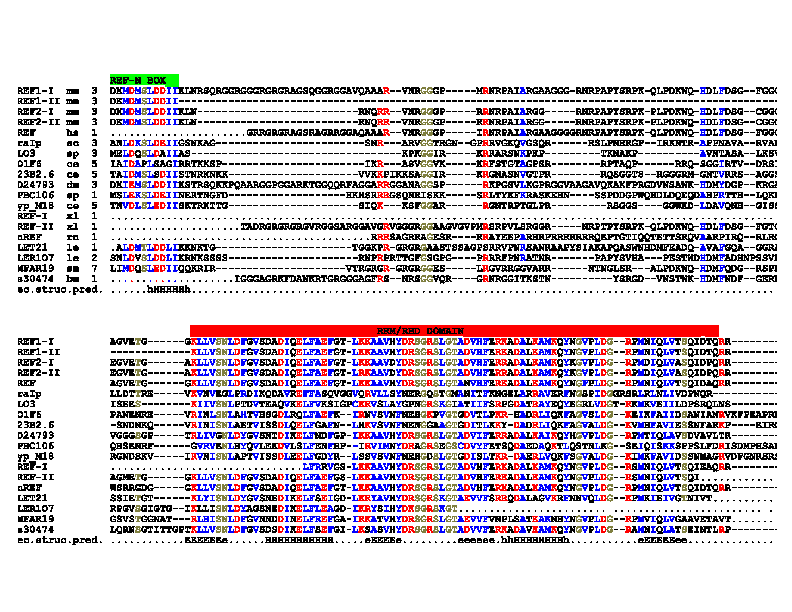
| Figure 2: REF: an evolutionarily conserved family of hnRNP-like proteins. Multiple alignment of members of the REF-family of proteins. First column: protein names; second column: species names (bm: Bombyx mori; ce: Caenorhabditis elegans; dm: Drosophila melanogaster; hs: Homo sapiens; le: Lycopersicon esculentum; mm: Mus musculus; rn: Rattus norvegicus; sc: Saccharomyces cerevisae; sm: Schistosoma mansoni; sp: Saccharomyces pombe; xl: Xenopus laevis); third column: start of sequences; right most column: database accession numbers. Conserved charged residues are shown in red; conserved hydrophobic residues are shown in blue; other conserved residues are shown in bold. Dots represent the lack of sequence information while dashes represent gaps in the alignment. This multiple alignment was constructed by ClustalX (Thompson, J. D.et al., 1997) and manually refined by SEAVIEW alignment editor (Galtier et al, 1996). The RRM/RBD-domain and the conserved REF-N and REF-C boxes flanking the RBD-domain are shown in colour above the sequences. The secondary structure prediction by PHD (Rost, B. et al., 1994) is indicated below the sequences (H,h: alpha-helix; E,e: beta-strand). Capital letters indicate prediction with expected average accuary >82%. The sequence data for mREFs 1 (I and II) and 2 (I and II) are available from GenBank/EMBL/DDBJ under accession numbers AJ252140 and AJ252141. |