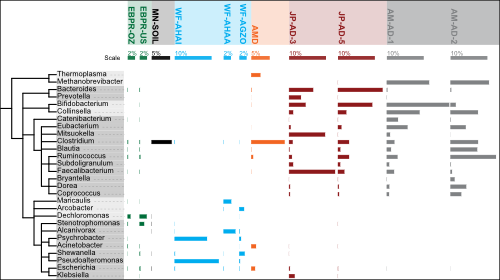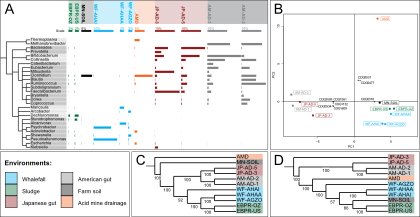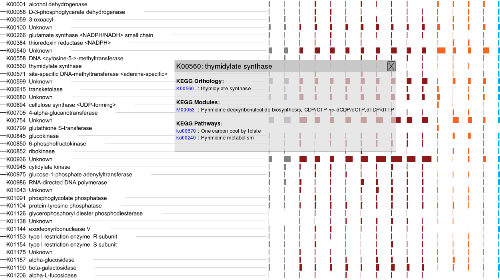
SmashCommunity v1.6 includes several months worth of improvements, bugfixes, new features and optimizations.
We know, there was never a v1.5, but that is because v1.5 was an internal release and it progressed so much
that we are at v1.6. Starting from v1.6, SmashCommunity supports functional annotation of metagenomes using
the KEGG functional database, which organizes genes at the orthologous group, functional module and functional
pathway levels. Check the image on the left to see the nature of functional information that you can obtain
using annotations with the KEGG database.
Minor update on Sep 09, 2011
There was a minor update to v1.6 on Sep 09 that fixes the URL for meta_rna/rRNA_hmm software, so that the installation
process does not stall. If you had trouble installing the older v1.6 package, please download the latest version.
The latest version is v1.6p2.
Note: KEGG functional database is not free anymore. We are working on ways to let the users download KEGG database file
by themselves and then install support for KEGG functional annotation. We hope to get this fixed by the end of
September 2011.
Please see
"Applying a patch" in Special Topics to learn how to upgrade to the newest patch.
Minor update on May 06, 2011
There was a minor update to v1.6 on May 06
- fixing a couple of bugs related to functional analysis,
- adding an easy option to use MySQL as the database of choice.
If you choose MySQL as the database of
choice, a MySQL database containing information about reference genomes and
reference proteins will also be downloaded now, which may take some time.
Please see
"Applying a patch" in Special Topics to learn how to upgrade to the newest patch.
Key Features in this version:
- functional annotation using the KEGG functional database
- ability to make your own reference genome database or reproduce what we make available (requires BioPerl to be installed)
- several new visualization of results:
- ability to visualize functional compositions of metagenomes on iTOL and browse through additional annotation information
- improved phylogenetic mapping of the microbial reference sequences to the RDP taxonomic tree by identifying the 16S rRNA gene sequences from them
- ability to analyze metagenomic sequences in the RDP taxonomic tree space and display the results on iTOL, either on the NCBI or on the RDP tree
- ability to analyze 16S sequences in the RDP taxonomic tree space and display the results on iTOL, either on the NCBI or on the RDP tree
Special thanks to:
- Ivica Letunic, EMBL/BioByte
- For making the coolest tree visualization tool,
iTOL, and for adding so
many features that SmashCommunity uses to display phylogenetic
and functional summaries of metagenomic samples,
- Takuji Yamada, EMBL
- For the KEGG information digest (proteins, KOs, modules, pathways) and the contribution to several quantitative procedures,
- Les Dethlefsen, Stanford
- For being a patient beta tester and providing valuable feedback in making v1.6 stable,
- Daniel R. Mende, EMBL
- For being the longest standing beta tester since Smash was actually in beta stage and the contribution to several quantitative prodecures,
- Jens Roat Kultima, EMBL
- For being a beta tester and finding errors in the website,
- Rest of Bork group, EMBL
- For moral support.
See the downloads section for older versions.